
Protect your brand with the most robust detection tools in the food industry
Veriflow® Listeria monocytogenes provides robust specificity and sensitivity across an extensive variety of environmental surfaces and multiple food and beverage matrices, without complex and time consuming sample preparation.
Veriflow technology is proven to provide rapid, accurate, actionable detection of pathogen and spoilage indicators – with no compromises on ease of use and affordability. The technology is easily deployed onsite allowing you to identify microbial risks, take corrective action, and prevent large-scale contamination.
The Veriflow system is comprised of a small customized thermocycler, pre-aliquoted PCR reagent tubes, and proprietary buffers and hand-held cassette. Test results are garnered with less than 15 minutes of end user hands-on time per sample.
Unmatched specificity
AOAC Performance Testing MethodsSM Program was utilized for validation and verification of assay performance, with no significant difference in performance compared to corresponding culture-based reference methods.
- Broad inclusivity of 50 Listeria monocytogenes serotypes with 100% detection rate
- Correctly excludes all applicable strains
Unrivaled sensitivity
The robust Veriflow platform performs at the highest levels of accuracy even in the most challenging matrices, with vastly simplified sample preparation.
- Target amplification of a conserved gene market for Listeria monocytogenes
- Successfully validated in Ready-to-Eat foods, milk and dairy product, nutraceuticals, confectionary goods and environmental surfaces
- Reliable results in even the most challenging matrices
Unsurpassed ease of use
The Veriflow system is ideally suited for onsite implementation, with a modest capital investment and affordable per test cost.
- Results in 24 hours versus 3-4 days with traditional methods
- Streamlines your quality management processes
- Easily implemented onsite with existing resources
- Eliminates the need for proprietary media, two-step enrichment, and DNA extraction sample preparation
Test protocol
ENRICH
After 18-24 hours enrichment, transfer 500 µl sample to provided 1.5 ml sampling tube. Heat sample for 10 minutes, then allow to cool to room temperature.

AMPLIFY
Transfer 5µl of heat inactivated sample into provided PCR reagent tube. Place tube into Veriflow Thermocycler and start 90 minute amplification cycle with one touch.
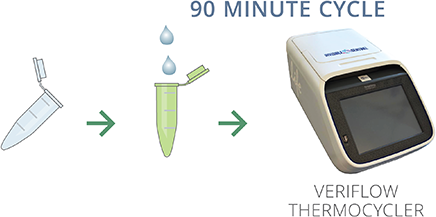
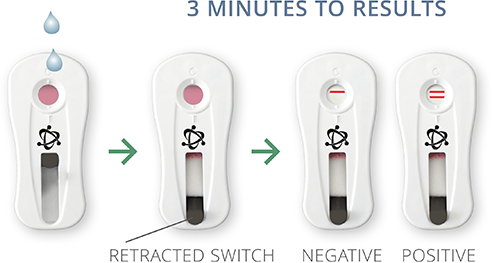
DETECT
Remove PCR reagent tube from thermocycler. Dispense contents of the tube onto test cassette window. Wait 3 minutes and retract test cassette switch to reveal test results. One line indicates a negative result, two lines indicate a positive result.
Technical specifications
Specificity
| Assay | Strains | Results | |
|---|---|---|---|
| Inclusivity | 50 Listeria monocytogenes Serotypes | 100% Detection Rate | Correctly identified all strains tested |
| Exclusivity | 39 Non-Listeria species | 100% Exclusion Rate | Correctly excluded all strains tested |
Sensitivity
| Matrix | Demonstrated equivalence to USDA MLG | External Validation | Client References |
|---|---|---|---|
| Stainless Steel | YES | YES | YES |
| Sealed Concrete | YES | YES | YES |
| Ceramic Tile | YES | YES | YES |
| Plastic | YES | YES | YES |
| Hot Dogs | YES | YES | YES |
| Deli Turkey | YES | YES | YES |
| Whey Powder | YES | YES | YES |
| 2% Pasteurized Milk | YES | YES | YES |
| Chocolate Chip Cookies | YES | YES | YES |
| Pea Protein | YES | YES | YES |
| Pea Fiber | YES | YES | YES |
| Rice Protein | YES | YES | YES |
| Fructose | YES | YES | YES |
| Chia | YES | YES | YES |
| Vitamin Mineral Pre-Mix | YES | YES | YES |
| Cocoa | YES | YES | YES |
Veriflow STEC Technical Specifications
| Specificity | 50 Listeria monocytogenes serotypes |
| Time to Results | 24 hours enrichment + 2 hour assay time |
| Matrix Compatibility | Wide variety of surfaces and matrices |
| Sensitivity | Zero tolerance detection |
| Assay Format | Qualitative |
| Test Stability | 1 year expiration with proper storage |
| Enrichment | Single step enrichment in Invisible Sentinel Listeria Enrichment Broth (ISLB) or mLEB No selective supplements required |
| Sample Preparation | Molecular platform that eliminates need for DNA extraction or purification |
| Workflow | Simple 3 step procedure |
| Results Interpretation | Immediate visualization on hand-held cassette - no complex data analysis |





